New protein-binding method may lead to reduction of drug side effects
By Steve KoppesNews Office
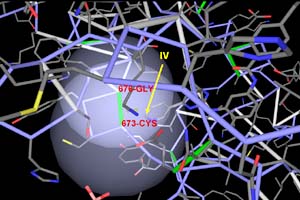 The light-colored oval on this computer-generated image of a protein highlights the location of a dehydron, an unprotected dry region where water can easily get in to destroy the hydrogen bond that holds proteins together. The colored bars designate various amino acids (two of which are labeled in red), the building blocks of proteins. The position of Gleevec, an anti-cancer drug, is labeled by the numeral IV. Scientists at the University and Rice University have shown that the distinctiveness of dehydrons can help biomedical researchers design drugs that are more effective by enabling them to distinguish between similar-looking proteins. | |
Scientists at the University and Rice University have successfully tested in the laboratory a new method for designing drugs that target specific disease-related proteins without creating undesirable side effects.
“Gleevec is the most popular cancer drug on the market, but it has side effects,” said Ariel Fernández, the Karl Hasselmann professor in bioengineering at Rice University in Houston. “This is essentially a new strategy to defeat the side effects.”
Fernández, along with Chicago professors Ridgway Scott and R. Stephen Berry, provided the details of their new drug-design method in the Tuesday, Jan. 10 issue of the Proceedings of the National Academy of Sciences. Clinical trials are not yet under way.
The team used its new method to redesign Gleevec, a drug with side effects that include peeling skin and ulcers. Some patients are so severely affected that treatment must be discontinued. “The particular drug in our tests is a cancer drug, but in principle the idea can be applied in other areas,” said Scott, the Louis Block Professor in Computer Science, Mathematics and the Physical Sciences Collegiate Division.
The new method exploits a sticky force that binds proteins, which are organic compounds essential to the function of living cells. The sticky force is associated with defects at the microscopic sites where hydrogen atoms bind proteins together. The scientists call these defects “dehydrons,” an idea they have developed in a series of papers published since 2002.
In a study published in 2003, Fernández and Scott showed that dehydrons signal the previously unpredictable location where proteins bind to one another. The following year, Fernández and Berry published another study explaining that, surprisingly, dehydrons displayed different structures in similar-looking proteins. The distinctiveness of dehydrons and their tendency to stick to oily molecules, the studies suggested, could be used to design inhibitor drugs that attach to specific proteins and prevent them from functioning.
“Cancer is a case of normal functions running out of control. So one way to stop cancer is to stop the normal functioning of a particular protein,” Scott said.
The Proceedings paper addresses a common problem with inhibitors, said Berry, the James Franck Distinguished Service Professor Emeritus in Chemistry and the College. “They are not always specific to the protein you want to stop, but sometimes stop other related proteins as well, proteins that the organism needs to keep functioning.”
The name “dehydron” reflects their preference to become dehydrated. Hydrogen bonds are wrapped by certain amino acids, the building blocks of proteins, to remain dry and strong. But dehydrons are exposed bonds where water can seep in and break apart the protein.
The Proceedings paper opens the possibility of treating diseases by designing drugs that wrap dehydrons. First, the drugs bind to the disease-related proteins by wrapping their dehydrons. Once bound, the drugs can then turn off the targeted proteins’ activities. Scott likens the action of the inhibitors to a key fitting into the tumblers of a lock. “If the key don’t fit, the protein won’t quit!” said Scott, paraphrasing the late Johnny Cochran.
This line of research began five years ago, after Fernández, then working in Argentina, spotted some of Berry’s papers on the structures of proteins. They began collaborating, which brought Fernández to the University’s Institute of Biophysical Dynamics for a year of sabbatical research.
Berry then introduced Fernández to Scott, who applies computers to the study of proteins. Together the three of them conducted a series of studies that led to the development of the dehydron concept and its application to drug design. “You’d have to call it a frontier idea at this point. It’s not in the textbooks,” said Berry of the dehydron.
In their latest research, the trio redesigned Gleevec, which fights cancer by suppressing the activity of certain proteins. But Gleevec, like other inhibitor drugs, sometimes affects both harmful proteins and also other untargeted proteins that have similar structures.
With funding provided by the National Institutes of Health, the team conducted laboratory experiments comparing the effects of Gleevec and an altered form of the drug on a protein associated with chronic myeloid leukemia. They found that the redesigned Gleevec more selectively affected the target protein while ignoring other proteins.
“The result is that you can design your inhibitor to fit just that one specific protein and not any old protein that’s in that same family,” Berry said. “It certainly is one of the ways you can minimize side effects.”
The new method is now being laboratory tested on cancer cells drawn from human patients through a separate collaboration of Fernández and colleagues at the at the M.D. Anderson Cancer Center in Houston. Initial results are encouraging and could lead to further testing in mice and then humans, Fernández said, adding, “There is a lot to be done.”
![[Chronicle]](/images/sidebar_header_oct06.gif)