Scientists identify defects in protein hydrogen bonds
By Steve KoppesNews Office
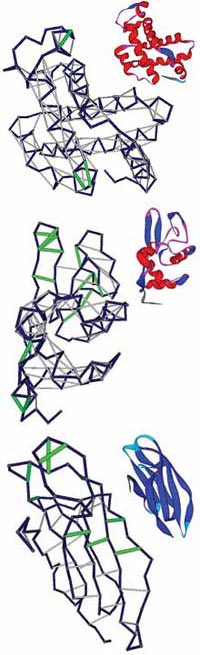 These illustrations show the pattern of underwrapped hydrogen bonds in the crystal structure of human apomyoglobin (which carries oxygen from the muscles), hen egg-white lysozyme (an enzymatic protein), and human microglobulin (an immune system protein). The ribbon representations are an aid to the eye. The protein backbones are presented in blue. The well-wrapped backbone hydrogen bonds show as grey segments, and the underwrapped hydrogen bonds are displayed as green segments. |
University scientists have discovered a new sticky force that binds together proteins, the stuff of which life is made. The discovery may lead to the development of drugs that prevent harmful proteins from attaching to one another.
“We believe this is a new force in nature,” said Ariel Fernández, a Visiting Scholar in the Institute for Biophysical Dynamics. “It’s never been properly characterized before and it seems to be at the core of biological phenomena when examined at the nanoscale.”
Fernández and Ridgway Scott, Professor in Computer Science and Mathematics, announced the discovery in the July 4 issue of Physical Review Letters.
“It’s a very radical way of thinking,” said Peter Rossky, the Marvin K. Collie-Welch regents chair in chemistry at the University of Texas, Austin. “This is an experiment which actually backs up that radical way of thinking, and that’s what’s striking about it.” The way proteins interact is central to biology, Fernández said. But scientists have long been confounded by their inability to predict exactly where proteins will bind to one another, even when their molecular structures are well-known.
“First of all, there’s almost nothing special about the binding site. It looks just like any other place on the protein,” Fernández said. “And secondly, when you look at the way they interact, it’s hardly ever a good match.”
Groups of atoms in a protein can either be hydrophobic (they do not interact with water), or nonhydrophobic (they do interact with water). One would expect that a hydrophobic group of atoms on a protein would bind with other hydrophobic groups. But instead, hydrophobic groups often will bind with nonhydrophobic groups.
Ironically, given the importance of water to the existence of life, Fernández and Scott found that the parts of proteins that would benefit most from staying dry were key to solving the puzzle.
“It turns out that water is not the friend of the protein. Water attacks proteins,” Scott said.
The attack occurs at the hydrogen bonds that hold proteins together. The building blocks of proteins are long strings of amino acids that fold up on themselves. In the process of folding, they make hydrogen bonds. Most of the hydrogen bonds get buried deep inside the protein, but some end up on the surface.
“Those are the ones that are vulnerable,” Scott said. “If you can break one apart, then you can attack deeper into the core and eventually break all of them apart. That’s part of how proteins get degraded.”
The hydrogen bonds of proteins must stay dry to remain strong. The strong ones have been wrapped in amino acid side chains that do not interact with water. These side chains are like strings of rope that wrap around the bond to keep them dry.
Under-wrapped or defective bonds are the ones most likely to become the sites where proteins will bond. In their Physical Review Letters paper, Fernández and Scott became the first scientists to measure the level of protection of hydrogen bonds in proteins.
“We identified packing defects, a hydrogen bond that is not properly wrapped,” Fernández said. In a separate paper that has been accepted for publication in the Biophysical Journal, he and Scott named the defect a “dehydron.”
“We call it a dehydron because it’s prone to being dehydrated,” Fernández said.
In the Physical Review Letters paper, Fernández and Scott identified dehydrons in six unrelated families of proteins that perform a variety of functions, from regulating the use of sugar to carrying oxygen from muscle. In all six families, they measured a comparable level of stickiness, which corresponds to a bond’s tendency to dehydrate.
“It couldn’t be by chance that they all gave the same result,” Fernández said. In their Biophysical Journal article, Fernández and Scott will report finding dehydrons at the binding sites of antibodies, the human immunodeficiency virus and the cold virus. “In everything we looked at, there was another example,” Scott said.
But finding the defective bonds solves only part of the puzzle. There remains the issue of how mismatched proteins bond together. Fernández and Scott addressed the issue as a three-body force rather than pairs.
“It’s not that A likes B or A likes C, but that A likes to dehydrate interactions when B is mated with C,” Fernández said.
The research provides insight into the molecular basis of cancer and amyloid-forming diseases such as Alzheimer’s. Such research may be of interest to drug companies, which often spend millions of dollars developing drugs that prevent binding over the entire surface of a protein.
The concept of the dehydron may make their task of developing inhibitor drugs easier. “Knowing where the binding sites are allows you to build drugs that will bind to that spot,” Scott said.
![[Chronicle]](/images/sidebar_header_oct06.gif)